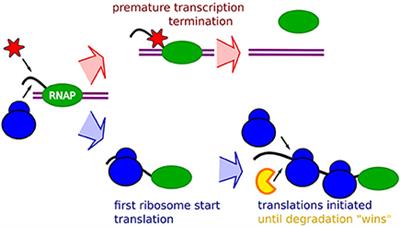
Frontiers | Occlusion of the Ribosome Binding Site Connects the Translational Initiation Frequency, mRNA Stability and Premature Transcription Termination
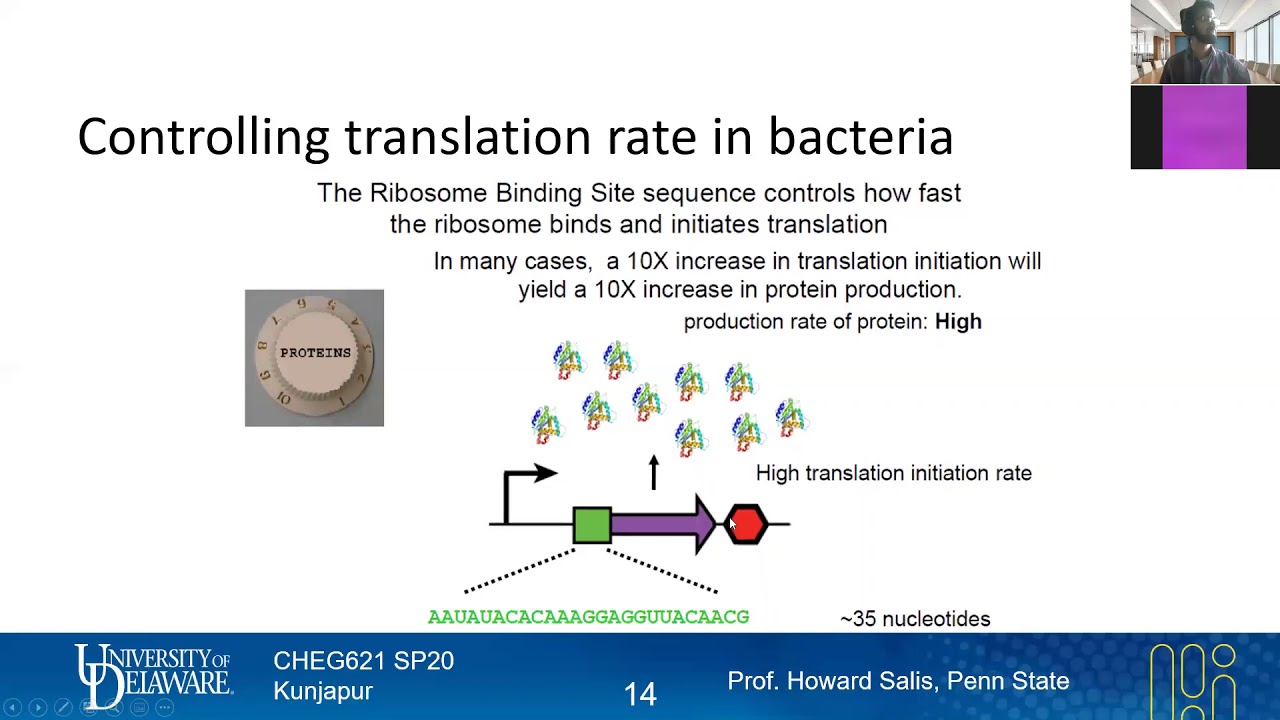
21 The Ribosome Binding Site | Expression Engineering | Lecture 14 | Metabolic Engineering | SP20 - YouTube
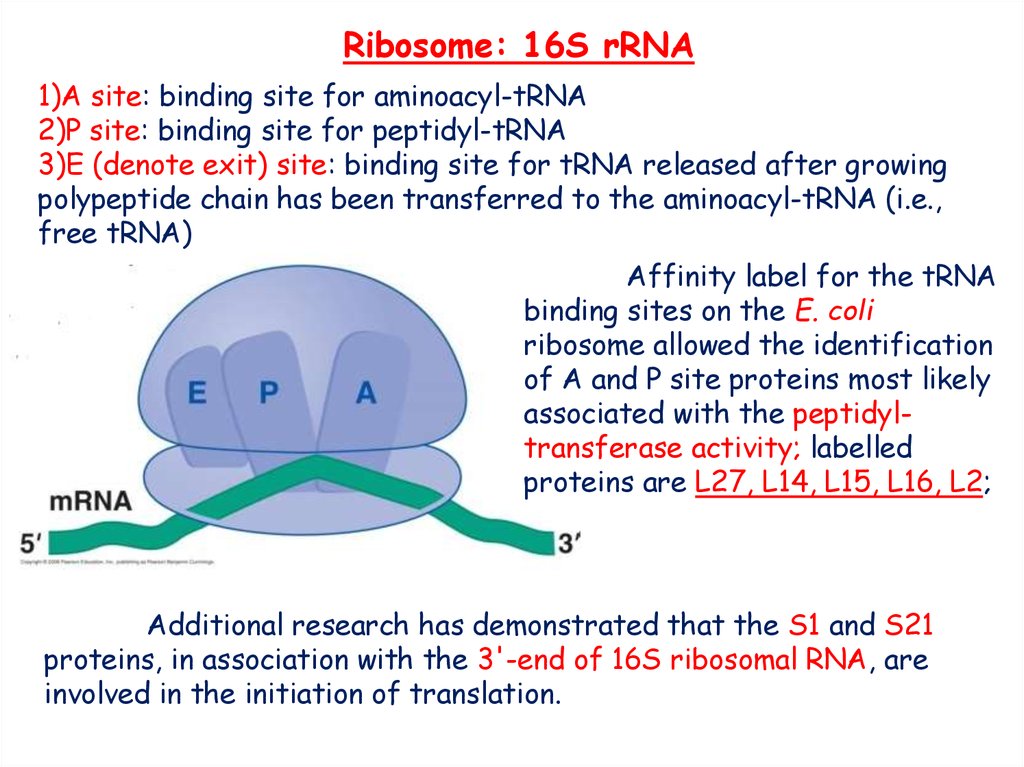
Initiation of translation in prokaryotes: initiation factors, initiator codons, 3'end of RNA small ribosomal subunit - презентация онлайн
Unusually Situated Binding Sites for Bacterial Transcription Factors Can Have Hidden Functionality | PLOS ONE
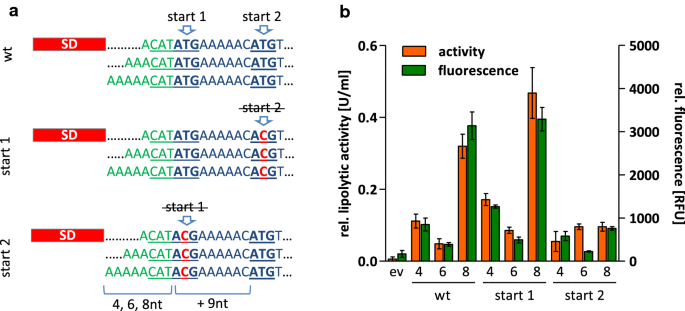
The length of ribosomal binding site spacer sequence controls the production yield for intracellular and secreted proteins by Bacillus subtilis | Microbial Cell Factories | Full Text
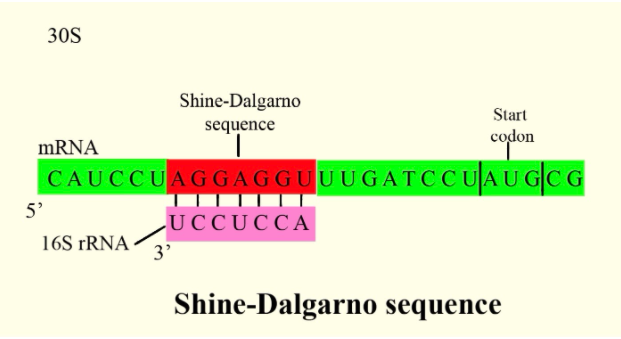
In prokaryotes, the ribosomes binding site on mRNA is called(a)Hogness sequence(b)Shine- Dalgarno sequence(c)Prinbow sequence(d)TATA- box

Determination of the ribosome-binding sequence and spacer length between binding site and initiation codon for efficient protein expression in Bifidobacterium longum 105-A - ScienceDirect
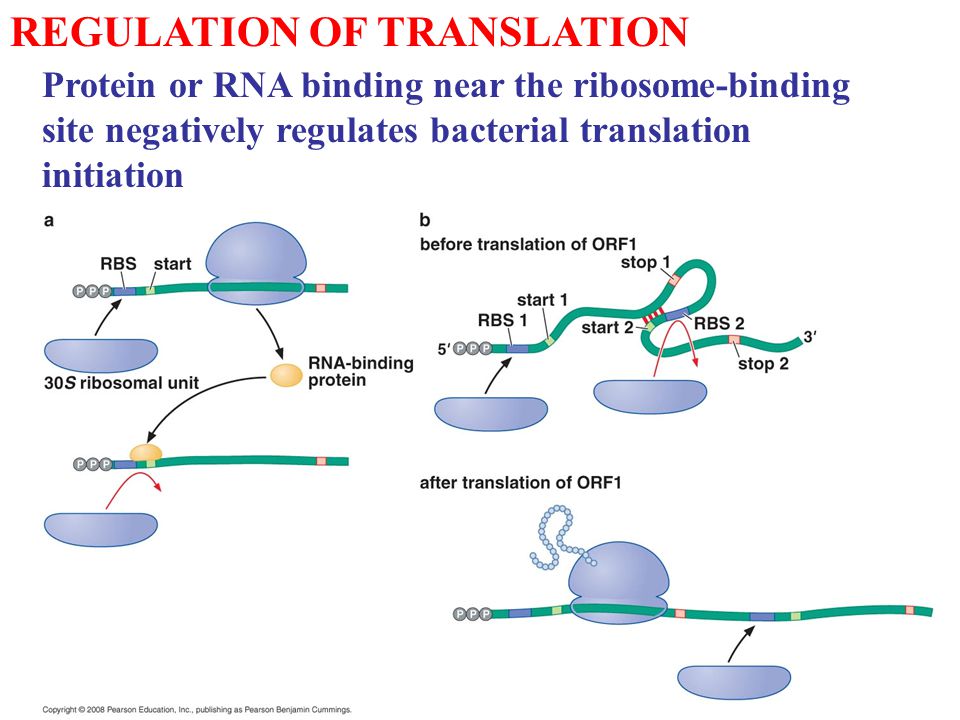
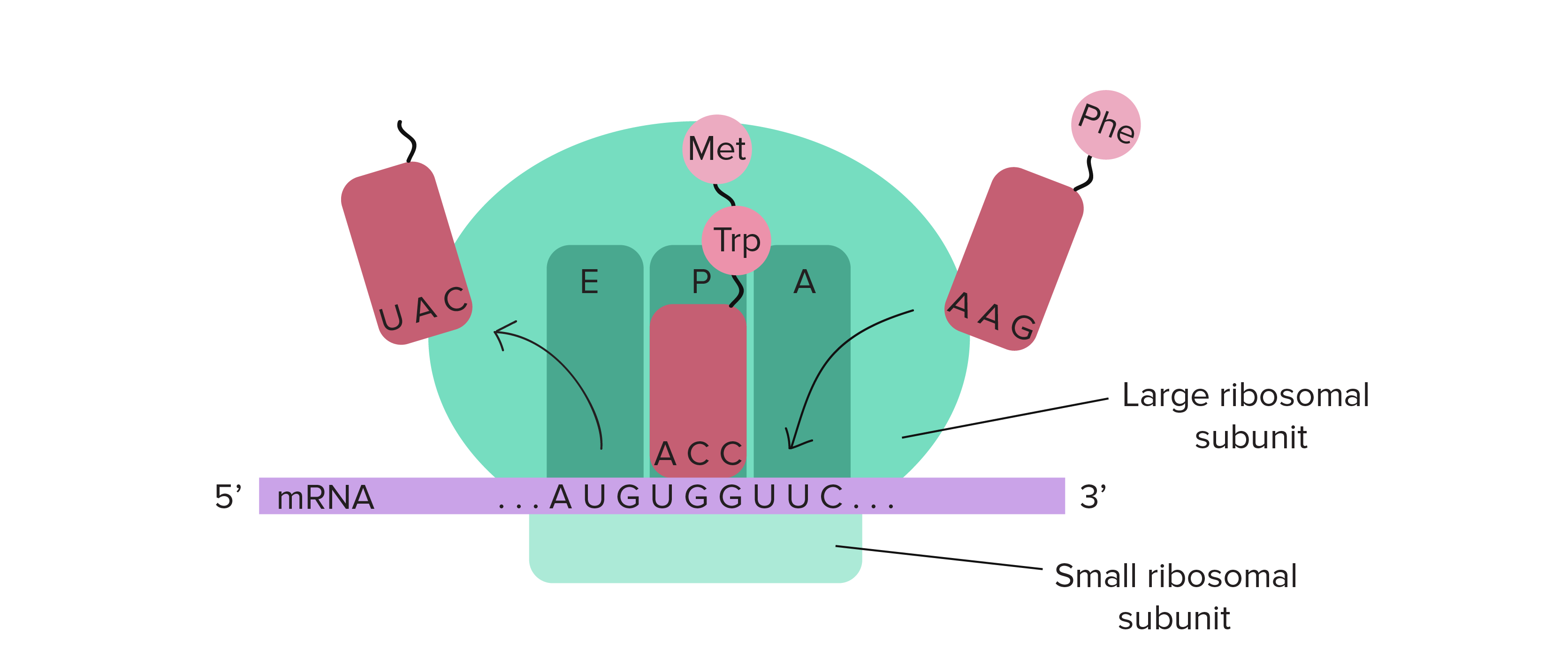
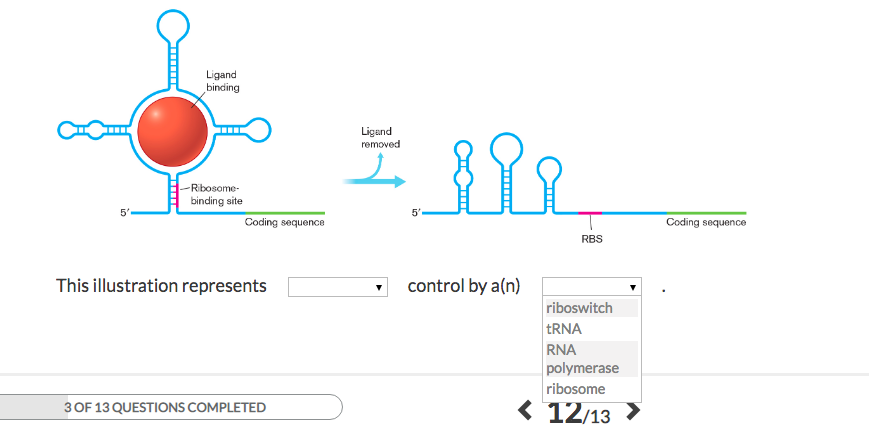
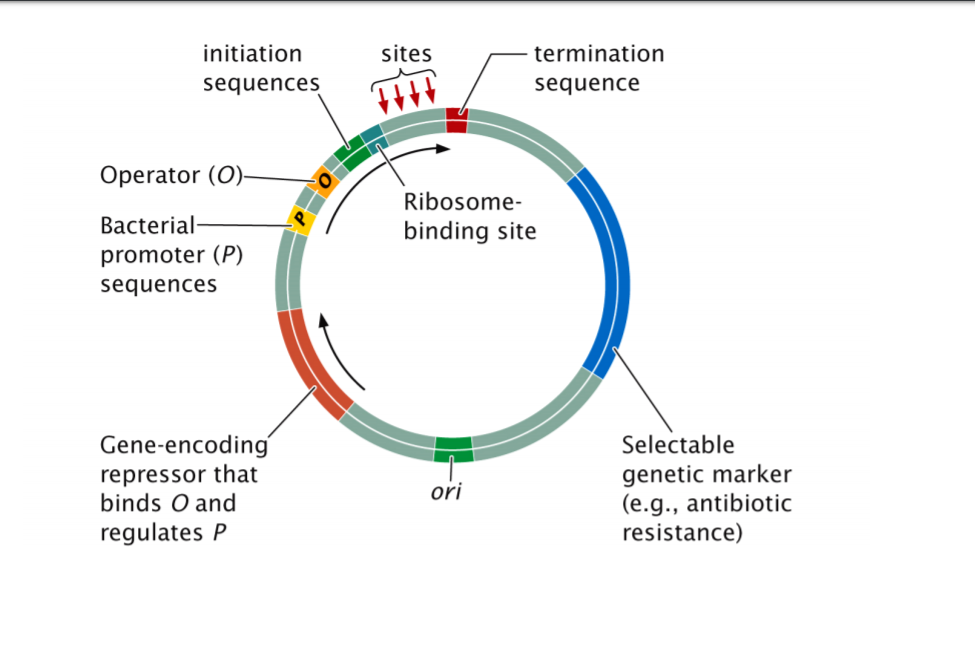
REGULATION OF TRANSLATION Protein or RNA binding near the ribosome-binding site negatively regulates bacterial translation initiation. - ppt download

Efficient engineering of chromosomal ribosome binding site libraries in mismatch repair proficient Escherichia coli | Scientific Reports
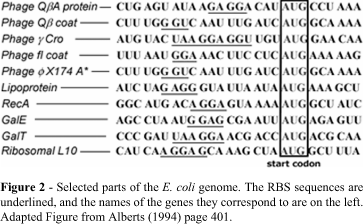
SciELO - Brasil - Ribosome binding site recognition using neural networks Ribosome binding site recognition using neural networks

The length of ribosomal binding site spacer sequence controls the production yield for intracellular and secreted proteins by Bacillus subtilis | Microbial Cell Factories | Full Text

The context of the ribosome binding site in mRNAs defines specificity of action of kasugamycin, an inhibitor of translation initiation | PNAS
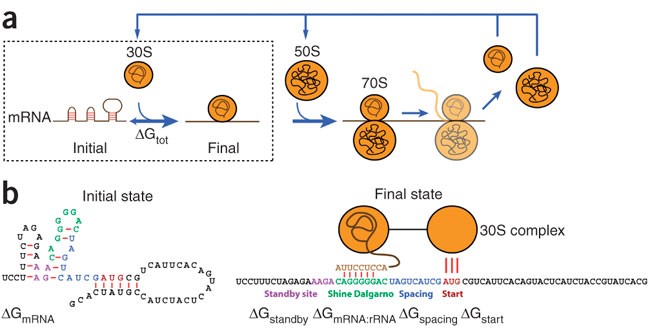
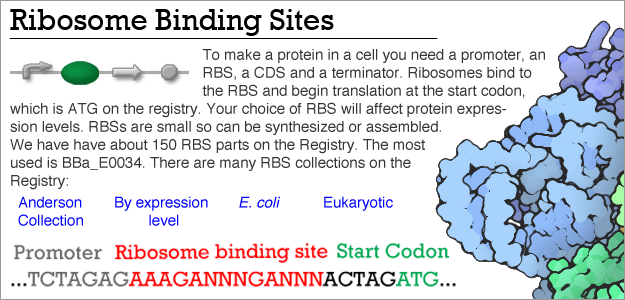
Automated design of synthetic ribosome binding sites to control protein expression | Nature Biotechnology

Sequence logos of putative promoters, ribosome-binding sites (RBS) and... | Download Scientific Diagram

1 The ribosome binding site and 5 0 protein coding sequence control an... | Download Scientific Diagram



![PDF] Classification of Escherichia coli K-12 ribosome binding sites | Semantic Scholar PDF] Classification of Escherichia coli K-12 ribosome binding sites | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/4af616123bce83b1f5dff449961c1df697c8d771/2-Figure2-1.png)