Long walk to genomics: History and current approaches to genome sequencing and assembly - ScienceDirect
Increasing calling accuracy, coverage, and read-depth in sequence data by the use of haplotype blocks | PLOS Genetics
Sequencing 101: ploidy, haplotypes, and phasing — how to get more from your sequencing data - PacBio
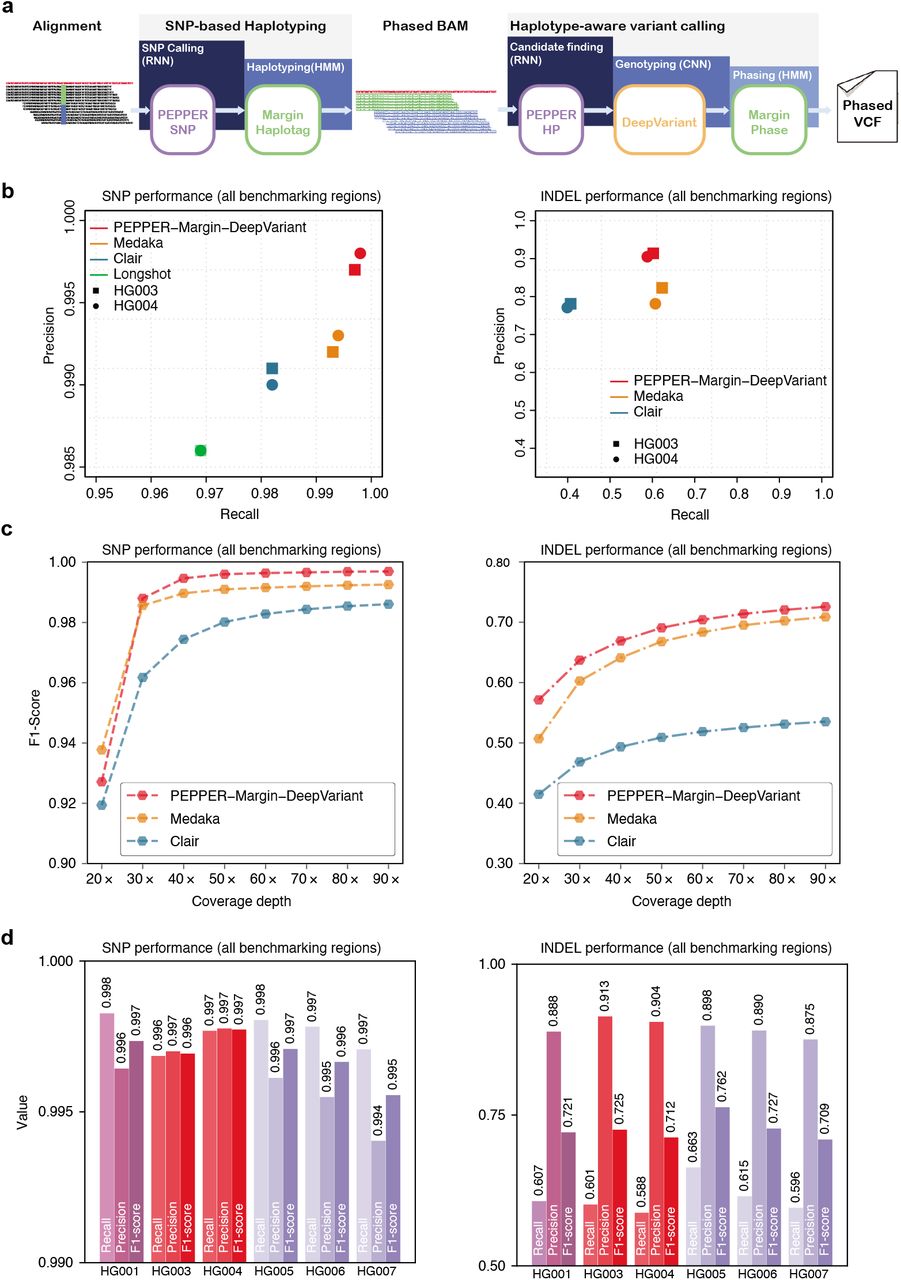
Haplotype-aware variant calling enables high accuracy in nanopore long-reads using deep neural networks | bioRxiv

Overview of the two main classes of haplotype phasing strategies. The... | Download Scientific Diagram
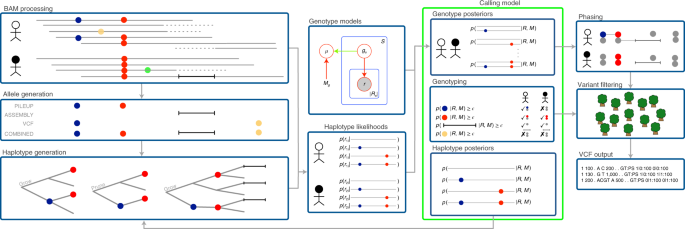
A unified haplotype-based method for accurate and comprehensive variant calling | Nature Biotechnology

Application of long-read sequencing to the detection of structural variants in human cancer genomes - ScienceDirect

Targeted linked-read sequencing for direct haplotype phasing of maternal DMD alleles: a practical and reliable method for noninvasive prenatal diagnosis | Scientific Reports
Sequencing 101: ploidy, haplotypes, and phasing — how to get more from your sequencing data - PacBio

Long-read sequencing of diagnosis and post-therapy medulloblastoma reveals complex rearrangement patterns and epigenetic signatures | bioRxiv
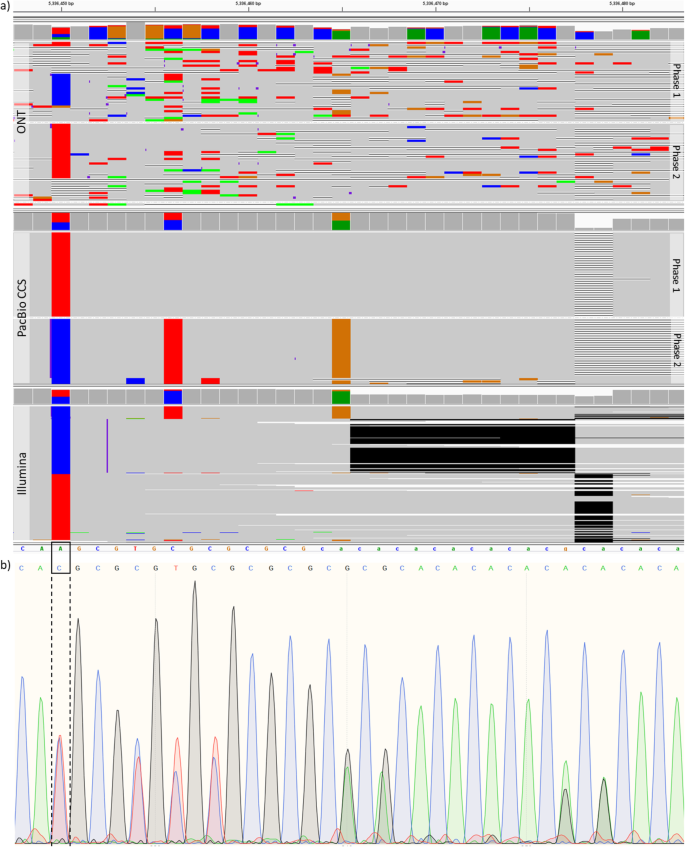
NanoCaller for accurate detection of SNPs and indels in difficult-to-map regions from long-read sequencing by haplotype-aware deep neural networks | Genome Biology | Full Text








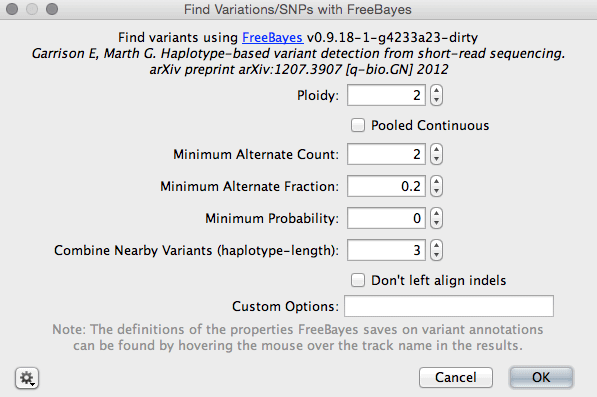
![PDF] Haplotype-based variant detection from short-read sequencing | Semantic Scholar PDF] Haplotype-based variant detection from short-read sequencing | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/cc5c9c18527d3f49f0a8c16308e4aa7c67f782ad/4-Figure1-1.png)
![PDF] Haplotype-based variant detection from short-read sequencing | Semantic Scholar PDF] Haplotype-based variant detection from short-read sequencing | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/cc5c9c18527d3f49f0a8c16308e4aa7c67f782ad/21-Figure6-1.png)